It is calculated by creating a distance matrix .
Hamming Distance is all about to find the number of differences in the alphabets at the same index of the compared strings . The number of difference in the position of alphabet would correspond to the number of substitutions required to make the strings equal .Here the alphabets are compared in CASE INSENSITIVE
,so, 'a' or 'A' is same here .
coding a game in java,programming a game in java,java game making,
for example here in this example we are comparing only the bases of the nucleic acids . There are four type of bases ,that is
'A' which stands for Adenine ,
'C' stands for Cytosine
'T' stands for Thymine
'G' stands for Guanine
So here we compare the strings which are made of the above mentioned alphabets .
Must Check: Snakes And Ladder Game
for example :
Let us consider two strings which consists only the bases of the nucleic acids ,
so suppose
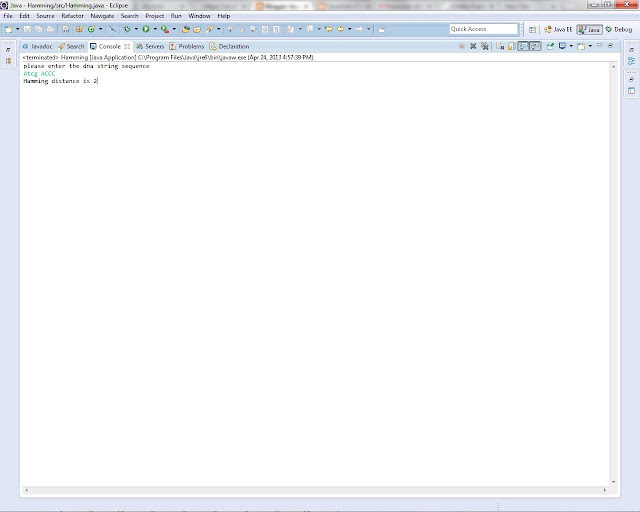
First string is "Atcg" , and
Second string is "ACCC"
coding a game in java,programming a game in java,java game making,
so here we can see at 1st position there is same alphabet 'a' . Moving forward to the next position there is 't'
in first string and 'C' in second , so these are clearly not equal so here is our first difference .
Similarly there is one more difference in last position of the both strings .So total there are two differences in the string , hence ,
Its hamming distance is 2 .
You can also calculate like this
A t c g
A C C C
----------------
0 1 0 1 , hence hamming distance is 2 .
----------------
So, we can also say that we are applying the XOR operation between two strings .The term hamming comes from the scientist Richard Hamming who first documented its details in the Hamming codes Error detecting and error correcting codes in 1950 . This algorithm has been applied in many fields in day to day life , one example of such field is telecommunication .
coding a game in java,programming a game in java,java game making,
Demo :
coding a game in java,programming a game in java,java game making,
import java.util.Scanner; public class Hamming { /** * @author Sonia * Hamming.class */ /** * @return true if and only if every character in the input String s is one of a, A, c, C, g, G, t or T. * @return false if s is null or empty. */ public static boolean isDNASequence(String s) { // TODO fill in this method if(s.length()==0 || s.equals(null)) return false; else { String temp= s.toUpperCase(); for (int i=0;i < temp.length();i++) { char c=temp.charAt(i); if( c=='A' || c=='T' || c== 'C' || c=='G' || c==' ') continue; else System.exit(0) ; } return true; } } /** * Get the distance matrix * * @param sequences - array of sequences * @return distance matrix */ public static int[][] getDistances(String[] sequences) { String s1=sequences[0]; String s2=sequences[1]; if(s1 == null || s2==null) System.exit(0); int m = s1.length(); int n = s2.length(); // normalize case s1 = s1.toUpperCase(); s2 = s2.toUpperCase(); int distance[][] =new int[40][40]; // Instead of a 2d array of space O(m*n) such as int d[][] = new int[m + // 1][n + 1], only the previous row and current row need to be stored at // any one time in prevD[] and currD[]. int prevD[] = new int[n + 1]; int currD[] = new int[n + 1]; int temp[]; // temporary pointer for swapping // the distance of any second string to an empty first string for (int j = 0; j < n + 1; j++) { prevD[j] = j; } // for each row in the distance matrix for (int i = 0; i < m; i++) { // the distance of any first string to an empty second string currD[0] = i + 1; char ch1 = s1.charAt(i); // for each column in the distance matrix for (int j = 1; j <= n; j++) { char ch2 = s2.charAt(j - 1); if (ch1 == ch2) { currD[j] = prevD[j - 1]; } else { currD[j] = Math.min(prevD[j] + 1, Math.min(currD[j - 1] + 1, prevD[j - 1] + 1)); } } temp = prevD; prevD = currD; currD = temp; } return distance; } /** * Get the hamming distance between two string sequences * * @param sequence1 - the first sequence * @param sequence2 - the second sequence * @return the hamming distance */ public static int getHammingDistance(String sequence1, String sequence2){ // TODO fill in this method int distance =0; if(sequence1 == null || sequence2==null) System.exit(0); sequence1 = sequence1.toUpperCase(); sequence2 = sequence2.toUpperCase(); if(sequence1.length() != sequence2.length()) { System.out.println("The string are not equal in length ,Please enter the strings wit equal lengths "); } for(int i=0;i < sequence1.length();i++) { if(sequence1.charAt(i)!=sequence2.charAt(i)) distance++; } return distance; } /** * Main method * * 1. Go through the parameters * 2. Ensure they are valid DNA sequences * 3. Get the hamming distance matrix * 4. Print out the highest difference * @param args - program arguments */ public static void main(String[] args) { // TODO fill in this main method code below this line System.out.println("please enter the dna string sequence"); char []store =new char[40]; String [] tokens=new String[40]; String [] subtoken=new String[40]; Scanner in =new Scanner(System.in); String s=in.nextLine(); isDNASequence(s); for(int i=0;i < s.length();i++) { store[i]=s.charAt(i); } tokens = s.split(" "); for(int m=0; m < tokens.length;m++){ subtoken[m]=tokens[m]; } getDistances(subtoken); int d= getHammingDistance(subtoken[0], subtoken[1]); System.out.print("Hamming distance is " + d); in.close(); } }